Research
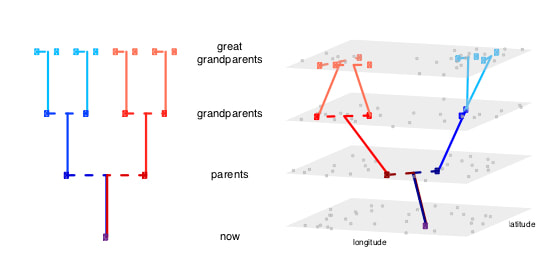 Put a pedigree in its geographical context and you can track the flow of ancestors across space through time.
Put a pedigree in its geographical context and you can track the flow of ancestors across space through time.
We've known for a long time that in many species, including humans, population genetic variation is distributed continuously across space, rather than partitioned between discrete groups. Historically, limitations in sampling have often allowed us to employ models of discrete structure. But, with the high-throughput genotyping and sequencing revolution and the massive empirical datasets it has facilitated (e.g., the 1000 Genomes project, the UK Biobank), we know we can no longer ignore geography. The failure to incorporate space into population genetic methods can have far-reaching consequences, from misleading inference of discrete population structure or admixture, to stratification issues in genotype-phenotype association studies, to a weaker basic knowledge of the biology of organisms. In the lab, we work to develop and implement novel population genetic models and statistical methods for describing population structure and admixture, as well as studying local adaptation, coevolution, and natural selection. This work combines population genetics theory, computation, statistics and inference, and a knowledge of the natural history of the empirical systems in which we apply our methods.
For more information on methods that have been developed and released, please visit the Methods page. For a full list of publications, see my Google Scholar profile.
For more information on methods that have been developed and released, please visit the Methods page. For a full list of publications, see my Google Scholar profile.