Gaia
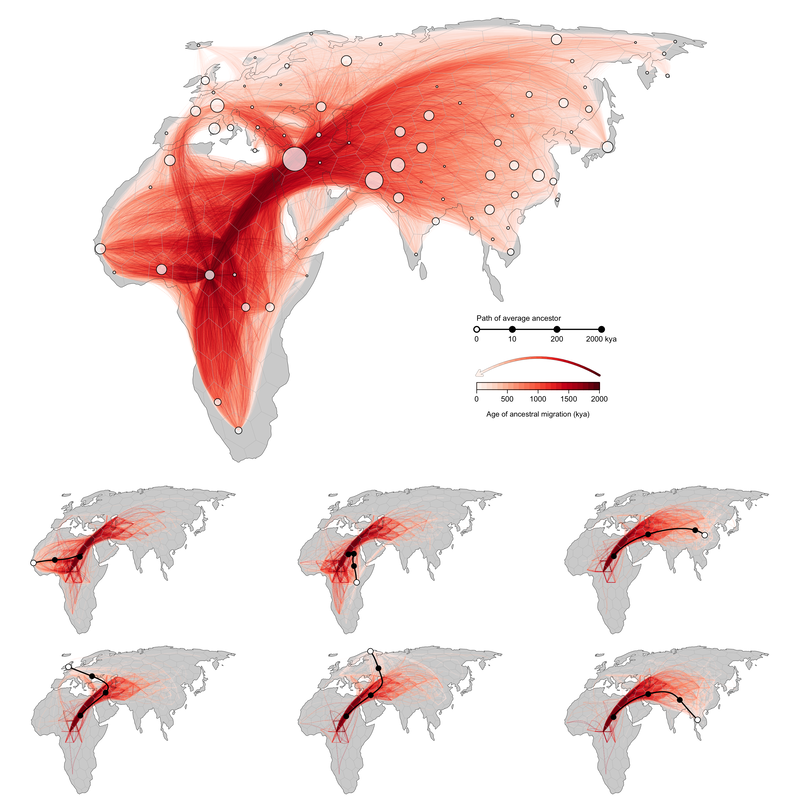
The genomes of present-day individuals existed at every point in the past, scattered across geographic space and contained within the ancestors from whom they will eventually inherit their genetic material. The way those ancestors moved across space through history determines spatial patterns of genetic relatedness in the present. If we knew the identities, locations, and ages of the ancestors of a sample, we could much more precisely and accurately report the geographic ancestry of a set of modern-day individuals through time. Moreover, we could learn about the history of dispersal in a species, identifying major population movements, demographic events, and barriers to migration.
We rarely have such detailed information, but recent advances in statistical and computational population genetics have facilitated the inference of an Ancestral Recombination Graph (ARG) from large numbers of whole genomes. The ARG is a record of all coalescence and recombination events since the divergence of the sequences under study, and therefore specifies a complete genealogy of the sample at each genomic position. This record can be represented as a tree sequence – an ordered set of trees, localized to adjacent regions of the genome, describing the gene genealogies of a set of samples at every genomic position. Each internal node in these local genealogies represents a haplotype within an ancestor from whom two or more sampled individuals have co-inherited a portion of their genome. By determining where and when each of these ancestors lived, we can, in principle, reconstruct the geographic history of a set of modern day individuals, documenting the path through space and time by which their genomes came to them.
Gaia is a new statistical method for inferring the geographic location of shared genetic ancestors in a tree sequence. It was developed by Mike Grundler (current postdoc in my lab) in collaboration with Jonathan Terhorst.
Instructions for how to install GAIA can be found on its github page here. The manuscript describing the method is on bioRxiv here.
We rarely have such detailed information, but recent advances in statistical and computational population genetics have facilitated the inference of an Ancestral Recombination Graph (ARG) from large numbers of whole genomes. The ARG is a record of all coalescence and recombination events since the divergence of the sequences under study, and therefore specifies a complete genealogy of the sample at each genomic position. This record can be represented as a tree sequence – an ordered set of trees, localized to adjacent regions of the genome, describing the gene genealogies of a set of samples at every genomic position. Each internal node in these local genealogies represents a haplotype within an ancestor from whom two or more sampled individuals have co-inherited a portion of their genome. By determining where and when each of these ancestors lived, we can, in principle, reconstruct the geographic history of a set of modern day individuals, documenting the path through space and time by which their genomes came to them.
Gaia is a new statistical method for inferring the geographic location of shared genetic ancestors in a tree sequence. It was developed by Mike Grundler (current postdoc in my lab) in collaboration with Jonathan Terhorst.
Instructions for how to install GAIA can be found on its github page here. The manuscript describing the method is on bioRxiv here.