Methods
GAIA
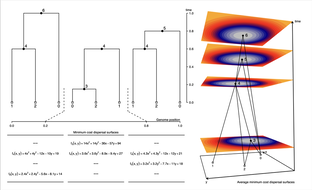
Gaia (for Geographic Ancestor Inference Algorithm) is a parsimony-based method that infers the geographic locations of shared genetic ancestors in a tree-sequence (the set of local gene genealogies across the genomes of a sequenced samples). Gaia works by fitting a minimum migration cost function to each genomic position in an ancestral haplotype using the generalized parsimony algorithm and the local gene genealogy relating the sampled genomes at the given position. By determining where and when each of these ancestors lived, we can, in principle, reconstruct the geographic history of a set of modern day individuals, documenting the path through space and time by which their genomes came to them.
Gaia (for Geographic Ancestor Inference Algorithm) is a parsimony-based method that infers the geographic locations of shared genetic ancestors in a tree-sequence (the set of local gene genealogies across the genomes of a sequenced samples). Gaia works by fitting a minimum migration cost function to each genomic position in an ancestral haplotype using the generalized parsimony algorithm and the local gene genealogy relating the sampled genomes at the given position. By determining where and when each of these ancestors lived, we can, in principle, reconstruct the geographic history of a set of modern day individuals, documenting the path through space and time by which their genomes came to them.
conStruct
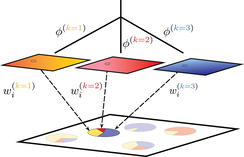
conStruct (for "continuous structure") is a model-based clustering algorithm (similar to STRUCTURE or ADMIXTURE) that models isolation by distance within populations. The method estimates ancestry proportions for each sample from a set of two-dimensional population layers, and, within each layer, estimates a rate at which relatedness decays with distance. This approach explicitly addresses the “clines versus clusters” problem in modeling population genetic variation. The method produces useful descriptions of structure in genetic relatedness in situations where separated, geographically distributed populations interact, as after a range expansion or secondary contact.
conStruct (for "continuous structure") is a model-based clustering algorithm (similar to STRUCTURE or ADMIXTURE) that models isolation by distance within populations. The method estimates ancestry proportions for each sample from a set of two-dimensional population layers, and, within each layer, estimates a rate at which relatedness decays with distance. This approach explicitly addresses the “clines versus clusters” problem in modeling population genetic variation. The method produces useful descriptions of structure in genetic relatedness in situations where separated, geographically distributed populations interact, as after a range expansion or secondary contact.
SpaceMix
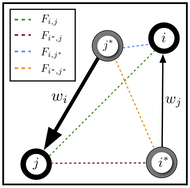
SpaceMix generates a “geogenetic” map of a genotyped individuals or populations. In the geogenetic map, distances between populations are effective distances, indicative of the way that populations perceive the distances between themselves. Nearby populations that are genetically dissimilar (e.g. separated by a barrier) are modeled as distant geogenetic locations, while two distant populations that are closely related (e.g., the parent and daughter populations of a recent expansion) are geogenetic neighbors. In this spatial context, “admixture” can be thought of as the outcome of unusually long-distance gene flow, which results in relatedness between populations that is anomalously high given the distance that separates them. This method depicts the effect of admixture using arrows, from a source of admixture to its target, on the inferred map. This geogenetic map is an intuitive and information-rich visual summary of patterns of population structure.
SpaceMix generates a “geogenetic” map of a genotyped individuals or populations. In the geogenetic map, distances between populations are effective distances, indicative of the way that populations perceive the distances between themselves. Nearby populations that are genetically dissimilar (e.g. separated by a barrier) are modeled as distant geogenetic locations, while two distant populations that are closely related (e.g., the parent and daughter populations of a recent expansion) are geogenetic neighbors. In this spatial context, “admixture” can be thought of as the outcome of unusually long-distance gene flow, which results in relatedness between populations that is anomalously high given the distance that separates them. This method depicts the effect of admixture using arrows, from a source of admixture to its target, on the inferred map. This geogenetic map is an intuitive and information-rich visual summary of patterns of population structure.

BEDASSLE
Estimates the relative contributions of pairwise geographic distance and pairwise ecological distance to patterns of genetic variation. Takes SNP data (in k populations at L loci) and pairwise geographic and ecological distances (between all k populations). Returns the effect sizes of these distances in a common currency. An example output is that 1000 meters difference in elevation is equal to ~200 kilometers pairwise geographic distance.
Estimates the relative contributions of pairwise geographic distance and pairwise ecological distance to patterns of genetic variation. Takes SNP data (in k populations at L loci) and pairwise geographic and ecological distances (between all k populations). Returns the effect sizes of these distances in a common currency. An example output is that 1000 meters difference in elevation is equal to ~200 kilometers pairwise geographic distance.